Nature Reviews Genetics@NatureRevGenet
Jul 13, 2022
6 tweets
Alternative splicing as a source of phenotypic diversity go.nature.com/3yW3Q3V #Review by
@Charlotte Wright, Christopher W. J. Smith & @Chris Jiggins
@Wellcome Sanger Institute @Darwin Tree of Life @Dept of Zoology @Cambridge Biochemistry @RNA labs
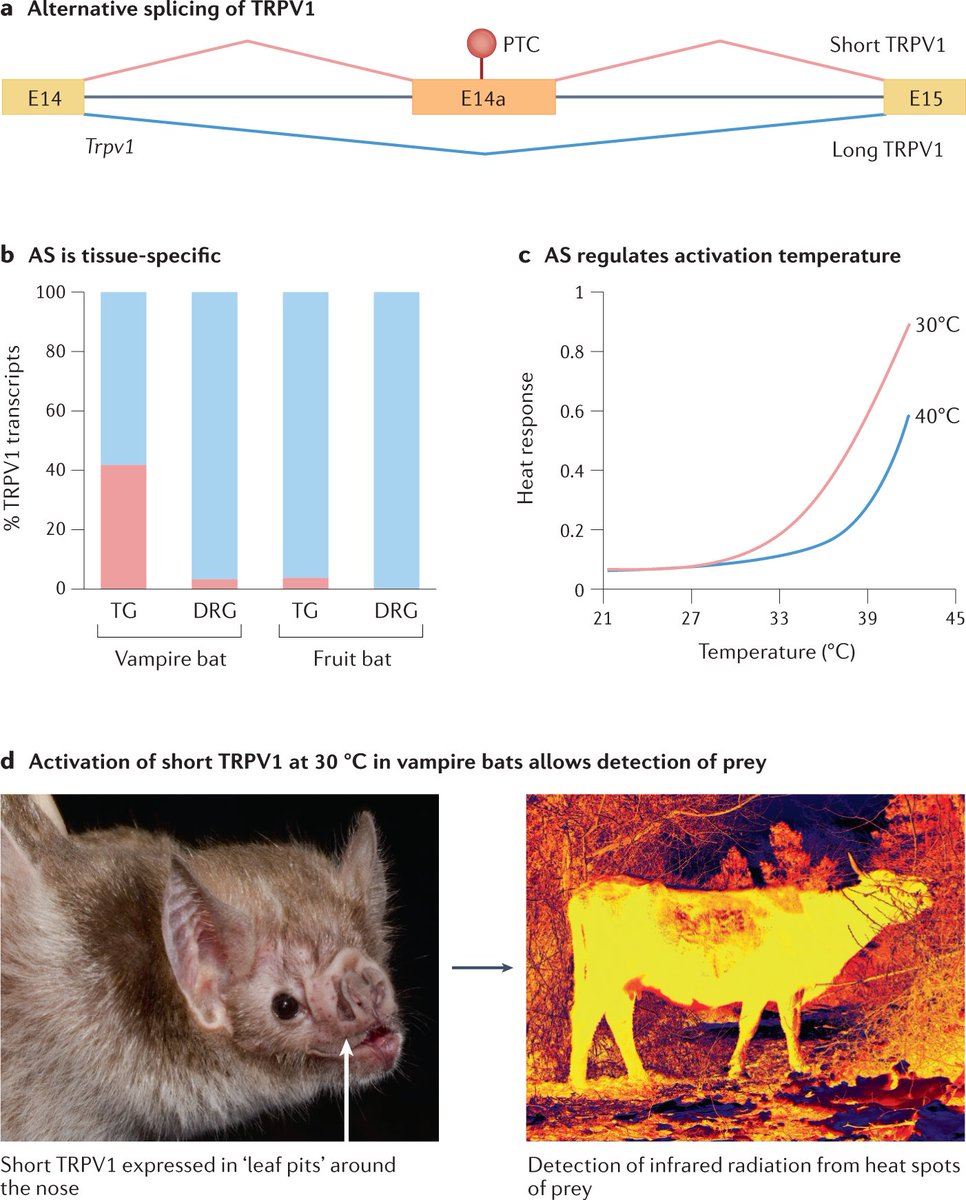
Alternative splicing generates multiple transcripts from a single gene, enriching the diversity of proteins and phenotypic traits. Alternative splicing contributes to key innovations over long evolutionary timescales, such as brain development in bilaterians.
Recent developments in long-read sequencing and high-quality genome assemblies for diverse organisms have facilitated comparisons of splicing profiles between closely related species, providing insights into how alternative splicing evolves over shorter timescales
In this Perspective, the authors summarize the ways in which alternative splicing occurs and the diverse functional roles it serves, namely in the context of proteomic diversity, development and phenotypic plasticity
They review the evolution of alternative splicing and the extent to which splicing variation can catalyse evolutionary change, including adaptation and speciation
Finally, the authors explore the evidence that alternative splicing diverges as a result of natural selection

Nature Reviews Genetics
@NatureRevGenet
Publishing reviews and commentaries across the fields of genetics and genomics. Part of @SpringerNature and @NaturePortfolio. Tweets are from the editors.
Missing some tweets in this thread? Or failed to load images or videos? You can try to .